Trained Immunity and Epigenetic Reprogramming
Molecular Biology Department, Radboud Institute of Molecular Life Sciences, Radboud and Radboud University Medical Center, Nijmegen, the Netherlands
Research topic
The term ‘epigenetics’ literally means ‘above DNA’ and refers to the study of molecular modifications that influence gene activity and chromosome structure. The two most commonly studied epigenetic processes are DNA methylation and histone modification, each of which is subject to developmental and environmental regulation and potentially modifiable with appropriate interventions.
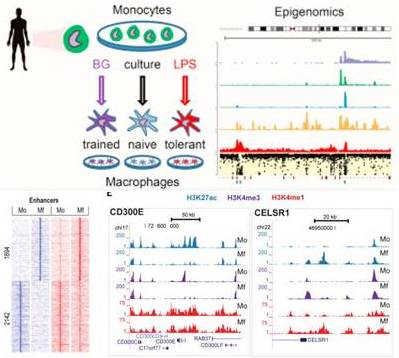
Accumulating evidence suggests that innate immune cells, such as monocytes, can be reprogrammed by exposure to microbe-associated molecular patterns (MAMPs) during their time in the circulation. This phenomenon is termed ‘innate immune memory’ and involves macrophage priming by exposure to microbe-associated molecular patterns (MAMPs) during their time as monocytes in the circulation or in their resident tissue. We have shown in several studies that tolerance (induced by E. colo lipopolysaccharide) and trained immunity (induced by C. albicans beta-glucan) are both associated with extensive epigenetic and transcriptional reprogramming. Notably, b-glucan induced trained immunity is associated with a unique repertoire of active enhancers, while LPS induced tolerance is associated with an inability to accumulate active histone marks at the promoter region. We have recently shown that ex vivo BG exposure can reverse both in vivo and ex vivo LPS-induced tolerance in monocytes, indicating that innate immune ‘trainers’ are good candidates for the treatment of diseases of the immune system.
Our current projects build on these pioneering studies and aim to increase our knowledge of innate immune memory. We apply state-of-the-art epigenomic, transcriptomic and gene-editing techniques to primary in vivo material, ex vivo stimulation experiments, and induced-pluripotent stem cells (iPS).
In collaboration with the Netea, Joosten and Riksen labs, we are also characterizing the epigenomic and transcriptomic signature of monocytes trained by metabolites such as fumarate, mevalonate, and uric acid, as well as the Bacillus Calmette–Guérin (BCG) vaccine, with the aim to identify core ‘pan-TRIM’ epigenetic signatures. Because epigenetic marks are reversible and modifiable by exogenous stimuli, they are prime targets for therapies looking to recover proper function in innate immune cells in disease.
Current projects of our group are:
- To catalogue changes in histone-modifications in human innate immune cells during monocyte-to-macrophage differentiation and establishment of trained innate immunity by different stimuli.
- To determine the role of key transcription factors in human monocyte differentiation and trained immunity, using both primary monocytes and induced pluripotent cells (iPS).
- To understand the mechanisms that control chromatin remodeling in endotoxin tolerance, using sepsis patient material, as well as in vivo and in vitro endotoxemia models
- To apply novel technology, such as scRNA-seq and low input ChIP-seq, to study innate immunity.
Group leaders:
- Henk Stunnenberg, PhD, professor in molecular biology
- Colin Logie, PhD, associate-professor in molecular biology
- Joost Martens, PhD, assistant-professor in molecular biology
Group members:
- Boris Novakovic, PhD, postdoctoral researcher
- Tessa van Kempen, PhD, postdoctoral researcher
- Shuangyin Wang, PhD student
- Ehsan Habibi, PhD student
- Farid Karamati, PhD student
- Nader Atlasi, PhD student
- Cheng Wang, PhD student
- Ruiqi Liu, PhD student
Selected Publications:
- Crisan T, et al. Uric acid priming in human monocytes is driven by the Akt-PRAS40 autophagy pathway. PNAS. 2017 114(21), 5485–5490.
- Bujko, A, Atlasy N et al, in revision
- Netea MG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016 Apr 22;352(6284):aaf1098
- Stunnenberg HG, et al. The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery. Cell. Nov 17;167(5)
- Novakovic B, et al. β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell. 2016 Nov 17;167(5):1354-1368.e14
- Arts RJ, et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 2016;24:807-819.
- Saeed S, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014 Sep 26;345(6204):1251086
- Cheng SC, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014 Sep 26;345(6204):1250684.
- Quintin J, et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012 Aug 16;12(2):223-32.
- Kleinnijenhuis J, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):17537-42
